

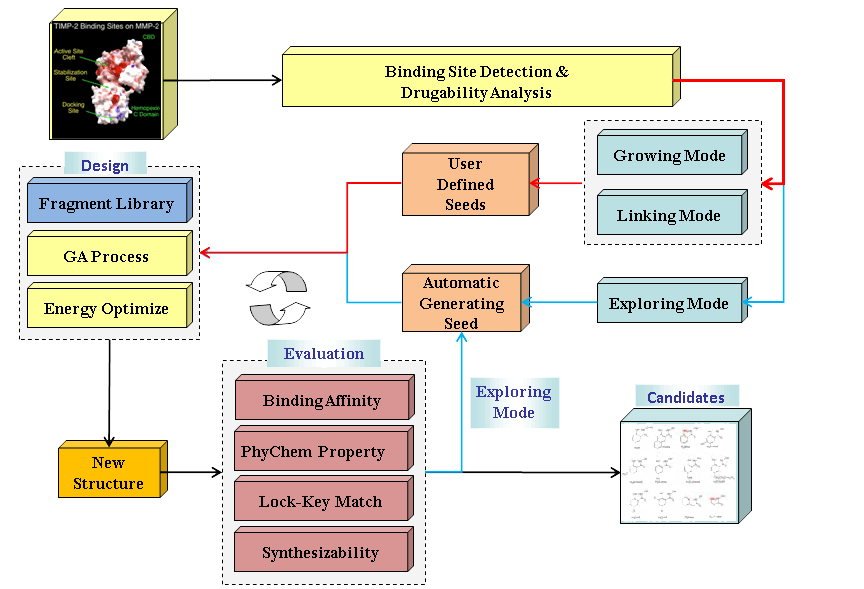
An overview of LigBuilder v2.0 program
Most input parameters are assembled into some default parameter sets, and users could simply adjust the design strategy by loading these sets. CAVITY:The default running settings file is under the path "LigBuilderV2/bin/cavity.input". BUILD:The default user settings file is under the path "LigBuilderV2/default/usersettings.input" and the default running settings file is in the path "LigBuilderV2/bin/build.input", please set the three files carefully before running your jobs.
The parameter file defines the input files, the output
files, and all the necessary parameters to run the program as you wish.
It is in plain text, in which each line starts with a key word. You can
find the detailed explanation of all the key words in the following pages
of this manual.
Example of parameter file:
POCKET_ATOM_FILE 1db4/1db4_pocket_1.txt //common
INCLUDE ../default/default.input //include
$NAME$ 1db4 //predefine (only valid in BUILD)
POCKET_GRID_FILE 1db4/$NAME$_grid_1.txt //common
[Extract]SEED_LIB 10000 //conditional (only valid in BUILD)
Common parameters are used for evaluation of the key words. Include parameter will guide the program to load another parameter sets. Predefined parameters will replace the corresponding string to the redefined string in all parameters inclulding these inherited from other parameter sets. In the example, the "POCKET_GRID_FILE" will be set to "1db4/1db4_grid_1.txt" for the string "$NAME$" has been replaced by string "1db4". Conditional parameters only take effect when the program executes the certain function or certain mode in the brackets. Attention: Predefine and Conditional parameters are just supported by BUILD.
CAVITY has three main functions: first, analyze the binding site and prepare the information necessary for running BUILD; second, estimation the ligandability of the binding site. Third, derive the key interaction sites within the binding site and suggest a pharmacophore model.
Synopsis of running CAVITY:
cavity Parameter_file
For example:
./cavity 1db4.input
The major function of BUILD is constructing ligand molecules for the target protein by applying the fragment-based design strategy. All the molecules are developed and evolved with a Genetic Algorithm procedure. All resultant molecules will be collected in a file and reported in HTML pages.
Synopsis of running BUILD:
build -Function Parameter_file [Id]
For example:
./build -Automatic build.input
Function List:
Automatic:
Automatic: Automatic design mode for one-stop drug design. With this mode, LigBuilder v2.0 will monitor all subroutines and accomplish all post-process procedure automatically. It is strongly recommend to run the design job with this mode.
Design:
Normal: Standard design mode. DO NOT access previous seed library. It will OVERWRITE previous result file.
Daemon: Daemon design mode, auto resume from faults. DO NOT access previous seed library. It will OVERWRITE previous result file.
Continue: Standard continue mode. DO access previous seed library. It will APPEND to previous result file.
DContinue:Daemon continue mode, auto resume from faults. DO access previous seed library. It will APPEND to previous result file.
Control:
PAUSE: Pause
CONTINUE: Continue
RELOAD: Reload the parameter file(only Daemon/DContinue mode)
EXIT: Exit
FINISH: Force to stop design process and come to the post-process.(only Automatic mode)
Process results:
Process: Convert result file (LIG format) to Mol2 format
Filter: Filtering results with structure constrains
YScore: Estimate the ligand binding affinity of results by YScore
Recommend: Help user to select top elites from large result sets.
Cluster: Clustering results
ReportCls: Report the clustering results in HTML
ReportSyn: Report the synthesis analysis results in HTML
Tools:
Extract: Extract seed structures from known inhibitors( for growing and linking strategies)
Results analysis:
Score: Estimate the ligand binding affinity of one molecule
Search: Searching the target substructure in the results.
Synthesize: Accurate synthesis analysis of results
Build database:
ExtLibrary: Build rigid fragments library ( extracting fragments from known inhibitors, for mimcs design)
RotLibrary: Build rotatable fragments library (extracting fragments from database)
SynDatabase: Build reaction database
MatDatabase: Build material database
1) Get receptor for RCSB PDB: Human non-pancreatic secretory phospholipase A2. (PDB entry 1db4):
receptor/1db4.pdb //PDB file of the protein (for CAVITY)
2) Prepare the input file for CAVITY
cavity.input // input the receptor path
-----------------------------------------------------------------
RECEPTOR_FILE receptor/1db4.pdb
-----------------------------------------------------------------
3) Run CAVITY:
./cavity cavity.input
4) Determine cavity id and estimate the ligandgability:
receptor/1db4_surface_1.pdb //
cavity id=1 and predicted maximal pKd=8.03
-----------------------------------------------------------------
REMARK 5 Pedicted Maximal pKd 8.03
-----------------------------------------------------------------
5) Prepare the input file for BUILD
build.input // input the receptor info from CAVITY
-----------------------------------------------------------------
POCKET_ATOM_FILE receptor/1db4_pocket_1.txt
POCKET_GRID_FILE receptor/1db4_grid_1.txt
-----------------------------------------------------------------
6) Run BUILD:
Start two shells first
Shell A:
./build -Automatic build.input
Shell B:
chmod +x run_1db4.list //you'd better run the list on your cluster rather
./run_1db4.list
//than on the system node in practice
NOTICE:
1: It is recommended to assign each task in the list file to a computer cluster rather than run them sequentially in practice.
2: It is not necessary to make all tasks run or finished. User can carry out tasks on different operation systems and then copy the output files to the outputting path assigned in the "LigBuilderV2/default/path.input". In this way, these tasks will NOT be monitored by the server, and please collect the output files before starting post-process.)
3: If a subroutine lose activity as appeared in the activity list, please check the memory status first and make sure that a suitable memory strategy is used. Try to terminate the subrountie and restart it. (Sometimes
the activity list would not capture the real status of subroutines,
especially when the harddisk or memory load is reaching the system
limit, that is, you'd better keep watching for a period of time to make
sure whether the subroutines are functional or not.)
4: Simply restart the server if it is abruptly terminated. The restarted server will reconnect to all active subroutines. DO NOT start a new server for one design task.
7) View the report pages when accomplished
result/report_1db4/report.html // report page index in HTML
result/report_1db4/images/ // directory of report pages and images
Sample parameter files, i.e. "build.input", "cavity.input", are also included in the same directory.
[Content] [Introduction] [Download] [Install] [Overview] [CAVITY] [BUILD] [Skills] [FAQs]
(These web pages are edited by Yaxia Yuan. Latest update: Jan. 2021)