Welcome to CorrSite, a server for the prediction of allosteric sites on protein structures.
CorrSite Server is a freely accessed web-server designed to identify potential protein allosteric sites. Here, we describe the CorrSite algorithm briefly. First, the program imports the coordinates of protein atoms from a PDB file and its orthosteric site information. Then, the program detects the potential protein binding sites by using CAVITY. The default parameters are used to detect cavities. More detail parameters could be set in this server (http://repharma.pku.edu.cn/cavity/). The calculated cavities with greater than 75% overlapping residues with the orthosteric site are excluded. After that, the program calculates correlations between these potential ligand-binding sites and corresponding orthosteric sites using a Gaussian network model (GNM). If the normalized correlation of one cavity is more than 0.5, this cavity is identified as a potential allosteric site.
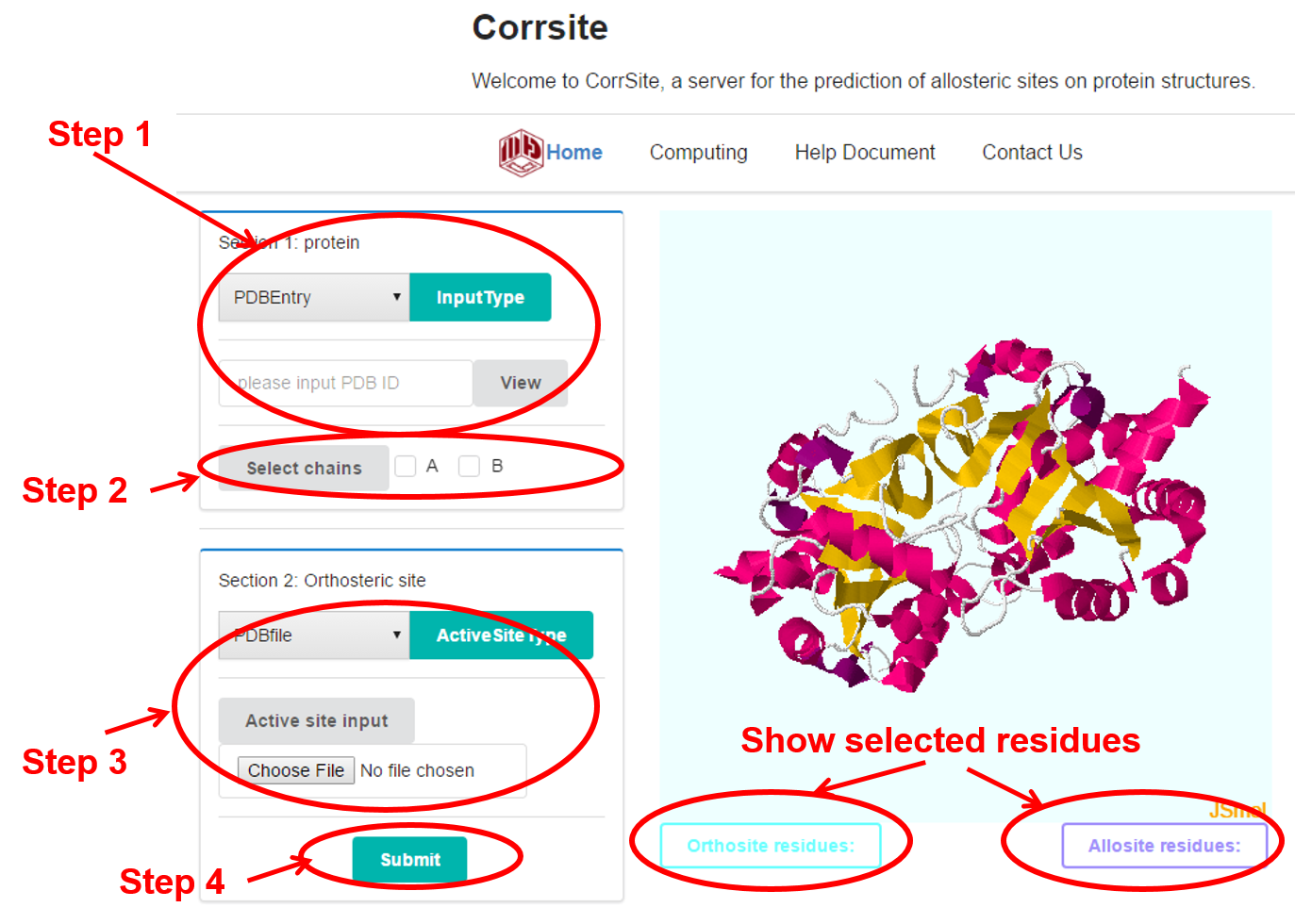
Load the protein of your interest. There are two provided ways: 1) load from RSCB based on JSmol; 2) Just manually upload the protein.
Select the chains of the whole protein. Note. Once the checkboxes were selected, the button of “select chains” must be clicked for generating the PDB file of the selected chains. The JSmol window will show the protein in cartoons;
Set the orthosteric sites:
46:A
47:A
49:A
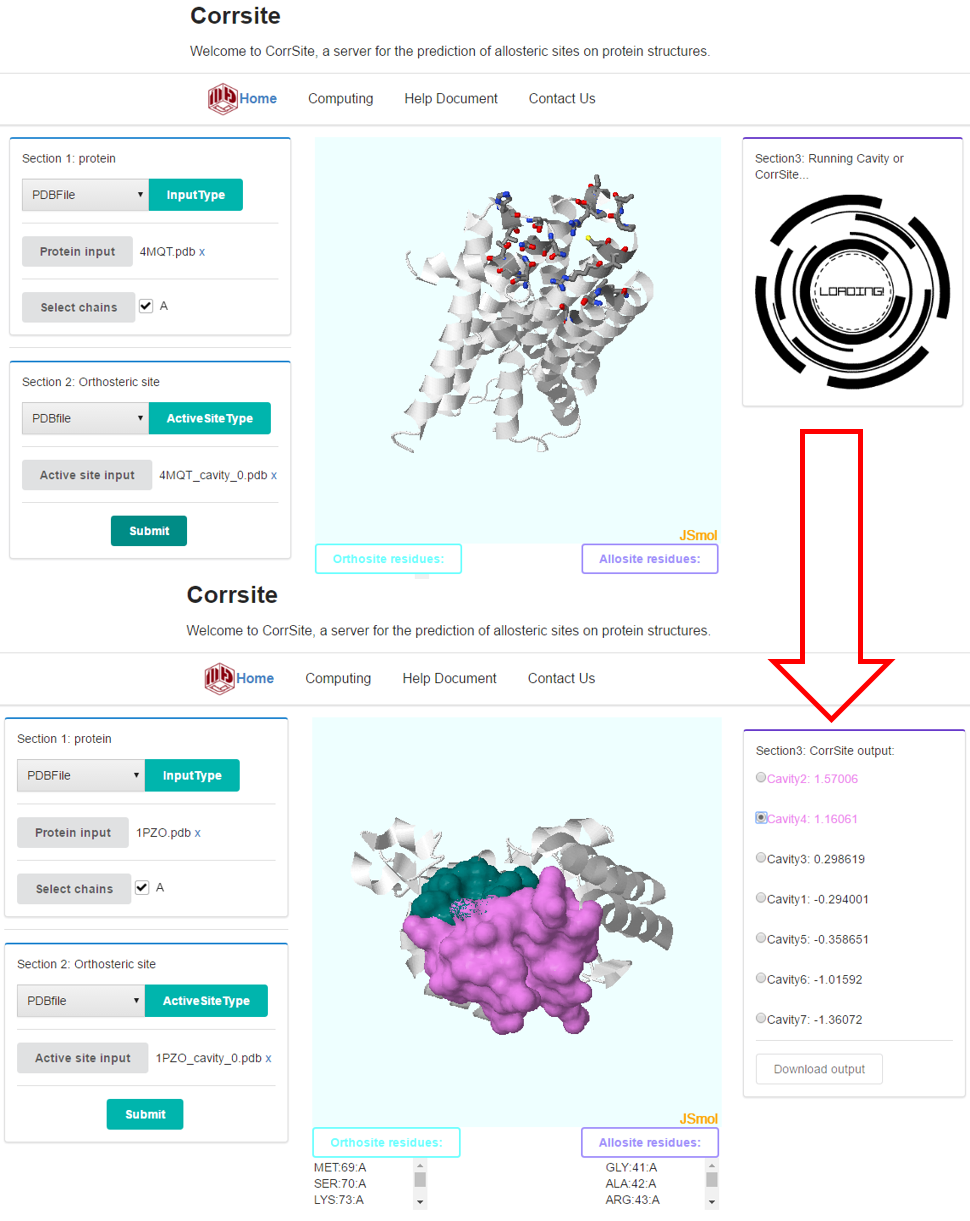
Click “Submit” button will submit your task. Then the Section 3 will come out. After finishing the task, Section 3 will output all the pockets and their scores, which measure the allosteric correlation of each pocket. The pocket with more than 0.5 is identified as a potential allosteric site. When the radio is checked, the JSmol window will show the surfaces of these residues in isoSurface solvent (radius 1.4Å).